Parameter of a Bivariate Copula for a given Kendall's Tau Value
Source:R/BiCopTau2Par.R
BiCopTau2Par.RdThis function computes the parameter of a (one parameter) bivariate copula for a given value of Kendall's tau.
BiCopTau2Par(family, tau, check.taus = TRUE)Arguments
- family
integer; single number or vector of size
n; defines the bivariate copula family:0= independence copula1= Gaussian copula2= Student t copula (Here only the first parameter can be computed)3= Clayton copula4= Gumbel copula5= Frank copula6= Joe copula13= rotated Clayton copula (180 degrees;survival Clayton'') \cr `14` = rotated Gumbel copula (180 degrees;survival Gumbel”)16= rotated Joe copula (180 degrees; “survival Joe”)23= rotated Clayton copula (90 degrees)
`24` = rotated Gumbel copula (90 degrees)
`26` = rotated Joe copula (90 degrees)
`33` = rotated Clayton copula (270 degrees)
`34` = rotated Gumbel copula (270 degrees)
`36` = rotated Joe copula (270 degrees)
Note that (with exception of the t-copula) two parameter bivariate copula families cannot be used.- tau
numeric; single number or vector of size
n; Kendall's tau value (vector with elements in \([-1,1]\)).- check.taus
logical; default is
TRUE; ifFALSE, checks for family/tau-consistency are omitted (should only be used with care).
Value
Parameter (vector) corresponding to the bivariate copula family and the value(s) of Kendall's tau (\(\tau\)).
No.
(family) | Parameter (par) |
1, 2 | \(\sin(\tau \frac{\pi}{2})\) |
3, 13 | \(2\frac{\tau}{1-\tau}\) |
4, 14 | \(\frac{1}{1-\tau}\) |
5 | no closed form expression (numerical inversion) |
6, 16 | no closed form expression (numerical inversion) |
23, 33 | \(2\frac{\tau}{1+\tau}\) |
24, 34 | \(-\frac{1}{1+\tau}\) |
26, 36 | no closed form expression (numerical inversion) |
Note
The number n can be chosen arbitrarily, but must agree across
arguments.
References
Joe, H. (1997). Multivariate Models and Dependence Concepts. Chapman and Hall, London.
Czado, C., U. Schepsmeier, and A. Min (2012). Maximum likelihood estimation of mixed C-vines with application to exchange rates. Statistical Modelling, 12(3), 229-255.
See also
Examples
## Example 1: Gaussian copula
tau0 <- 0.5
rho <- BiCopTau2Par(family = 1, tau = tau0)
BiCop(1, tau = tau0)$par # alternative
#> [1] 0.7071068
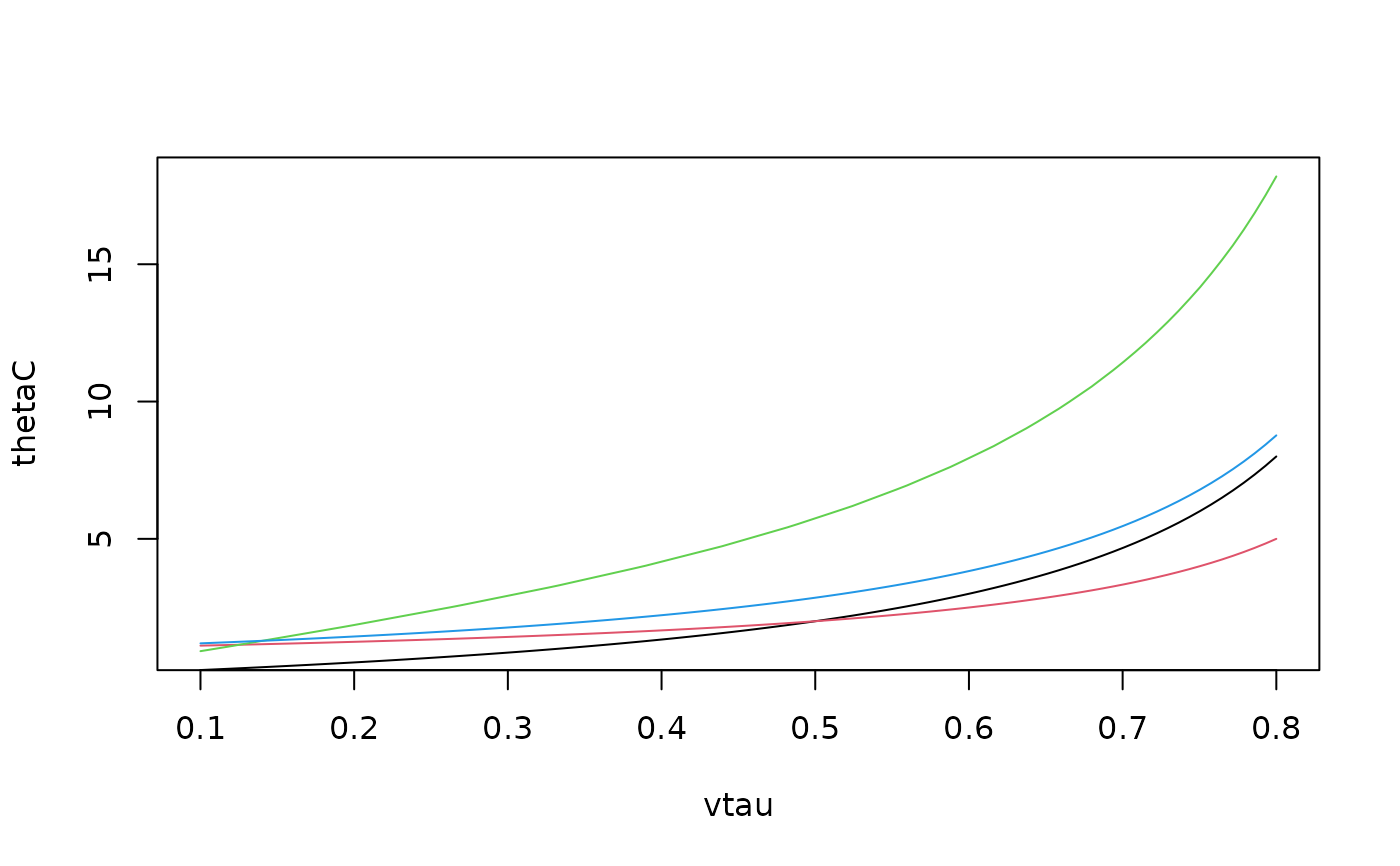
## Example 2:
vtau <- seq(from = 0.1, to = 0.8, length.out = 100)
thetaC <- BiCopTau2Par(family = 3, tau = vtau)
thetaG <- BiCopTau2Par(family = 4, tau = vtau)
thetaF <- BiCopTau2Par(family = 5, tau = vtau)
thetaJ <- BiCopTau2Par(family = 6, tau = vtau)
plot(thetaC ~ vtau, type = "l", ylim = range(thetaF))
lines(thetaG ~ vtau, col = 2)
lines(thetaF ~ vtau, col = 3)
lines(thetaJ ~ vtau, col = 4)
 ## Example 3: different copula families
theta <- BiCopTau2Par(family = c(3,4,6), tau = c(0.4, 0.5, 0.6))
BiCopPar2Tau(family = c(3,4,6), par = theta)
#> [1] 0.4 0.5 0.6
## Example 3: different copula families
theta <- BiCopTau2Par(family = c(3,4,6), tau = c(0.4, 0.5, 0.6))
BiCopPar2Tau(family = c(3,4,6), par = theta)
#> [1] 0.4 0.5 0.6