Sequential Pair-Copula Selection and Estimation for R-Vine Copula Models
Source:R/RVineCopSelect.R
RVineCopSelect.RdThis function fits a R-vine copula model to a d-dimensional copula data set.
Pair-copula families are selected using BiCopSelect() and
estimated sequentially.
RVineCopSelect(
data,
familyset = NA,
Matrix,
selectioncrit = "AIC",
indeptest = FALSE,
level = 0.05,
trunclevel = NA,
weights = NA,
rotations = TRUE,
se = FALSE,
presel = TRUE,
method = "mle",
cores = 1
)Arguments
- data
N x d data matrix (with uniform margins).
- familyset
integer vector of pair-copula families to select from. The vector has to include at least one pair-copula family that allows for positive and one that allows for negative dependence. Not listed copula families might be included to better handle limit cases. If
familyset = NA(default), selection among all possible families is performed. If a vector of negative numbers is provided, selection among all butabs(familyset)is performed. Coding of pair copula families is the same as inBiCop().- Matrix
lower or upper triangular d x d matrix that defines the R-vine tree structure.
- selectioncrit
Character indicating the criterion for pair-copula selection. Possible choices:
selectioncrit = "AIC"(default),"BIC", or"logLik"(seeBiCopSelect()).- indeptest
Logical; whether a hypothesis test for the independence of
u1andu2is performed before bivariate copula selection (default:indeptest = FALSE; seeBiCopIndTest()). The independence copula is chosen for a (conditional) pair if the null hypothesis of independence cannot be rejected.- level
numeric; significance level of the independence test (default:
level = 0.05).- trunclevel
integer; level of truncation.
- weights
Numerical; weights for each observation (optional).
- rotations
logical; if
TRUE, all rotations of the families infamilysetare included.- se
Logical; whether standard errors are estimated (default:
se = FALSE).- presel
Logical; whether to exclude families before fitting based on symmetry properties of the data. Makes the selection about 30\ (on average), but may yield slightly worse results in few special cases.
- method
indicates the estimation method: either maximum likelihood estimation (
method = "mle"; default) or inversion of Kendall's tau (method = "itau"). Formethod = "itau"only one parameter families and the Student t copula can be used (family = 1,2,3,4,5,6,13,14,16,23,24,26,33,34or36). For the t-copula,par2is found by a crude profile likelihood optimization over the interval (2, 10].- cores
integer; if
cores > 1, estimation will be parallelized within each tree (usingparallel::parLapply()). Note that parallelization causes substantial overhead and may be slower than single-threaded computation when dimension, sample size, or family set are small ormethod = "itau".
Value
An RVineMatrix() object with the selected families
(RVM$family) as well as sequentially
estimated parameters stored in RVM$par and RVM$par2. The object
is augmented by the following information about the fit:
- se, se2
standard errors for the parameter estimates (if
se = TRUE; note that these are only approximate since they do not account for the sequential nature of the estimation,- nobs
number of observations,
- logLik, pair.logLik
log likelihood (overall and pairwise)
- AIC, pair.AIC
Akaike's Informaton Criterion (overall and pairwise),
- BIC, pair.BIC
Bayesian's Informaton Criterion (overall and pairwise),
- emptau
matrix of empirical values of Kendall's tau,
- p.value.indeptest
matrix of p-values of the independence test.
#'
Details
R-vine copula models with unknown structure can be specified using
RVineStructureSelect().
Note
For a comprehensive summary of the vine copula model, use
summary(object); to see all its contents, use str(object).
References
Brechmann, E. C., C. Czado, and K. Aas (2012). Truncated regular vines in high dimensions with applications to financial data. Canadian Journal of Statistics 40 (1), 68-85.
Dissmann, J. F., E. C. Brechmann, C. Czado, and D. Kurowicka (2013). Selecting and estimating regular vine copulae and application to financial returns. Computational Statistics & Data Analysis, 59 (1), 52-69.
Examples
# define 5-dimensional R-vine tree structure matrix
Matrix <- c(5, 2, 3, 1, 4,
0, 2, 3, 4, 1,
0, 0, 3, 4, 1,
0, 0, 0, 4, 1,
0, 0, 0, 0, 1)
Matrix <- matrix(Matrix, 5, 5)
# define R-vine pair-copula family matrix
family <- c(0, 1, 3, 4, 4,
0, 0, 3, 4, 1,
0, 0, 0, 4, 1,
0, 0, 0, 0, 3,
0, 0, 0, 0, 0)
family <- matrix(family, 5, 5)
# define R-vine pair-copula parameter matrix
par <- c(0, 0.2, 0.9, 1.5, 3.9,
0, 0, 1.1, 1.6, 0.9,
0, 0, 0, 1.9, 0.5,
0, 0, 0, 0, 4.8,
0, 0, 0, 0, 0)
par <- matrix(par, 5, 5)
# define second R-vine pair-copula parameter matrix
par2 <- matrix(0, 5, 5)
## define RVineMatrix object
RVM <- RVineMatrix(Matrix = Matrix, family = family,
par = par, par2 = par2,
names = c("V1", "V2", "V3", "V4", "V5"))
## simulate a sample of size 500 from the R-vine copula model
set.seed(123)
simdata <- RVineSim(500, RVM)
## determine the pair-copula families and parameters
RVM1 <- RVineCopSelect(simdata, familyset = c(1, 3, 4, 5 ,6), Matrix)
## see the object's content or a summary
str(RVM1)
#> List of 20
#> $ Matrix : num [1:5, 1:5] 5 2 3 1 4 0 2 3 4 1 ...
#> $ family : num [1:5, 1:5] 0 1 3 4 4 0 0 3 4 1 ...
#> $ par : num [1:5, 1:5] 0 0.211 0.906 1.495 3.895 ...
#> $ par2 : num [1:5, 1:5] 0 0 0 0 0 0 0 0 0 0 ...
#> $ names : chr [1:5] "V1" "V2" "V3" "V4" ...
#> $ MaxMat : num [1:5, 1:5] 5 2 3 4 4 0 2 3 4 1 ...
#> $ CondDistr :List of 2
#> ..$ direct : logi [1:5, 1:5] FALSE TRUE TRUE TRUE TRUE FALSE ...
#> ..$ indirect: logi [1:5, 1:5] FALSE FALSE FALSE FALSE FALSE FALSE ...
#> $ type : chr "R-vine"
#> $ tau : num [1:5, 1:5] 0 0.135 0.312 0.331 0.743 ...
#> $ taildep :List of 2
#> ..$ upper: num [1:5, 1:5] 0 0 0 0.41 0.805 ...
#> ..$ lower: num [1:5, 1:5] 0 0 0.465 0 0 ...
#> $ beta : num [1:5, 1:5] 0 0.135 0.311 0.329 0.747 ...
#> $ nobs : int 500
#> $ logLik : num 2036
#> $ pair.logLik : num [1:5, 1:5] 0 10.5 83.5 87.1 487 ...
#> $ AIC : num -4052
#> $ pair.AIC : num(0)
#> $ BIC : num -4010
#> $ pair.BIC : num(0)
#> $ emptau : num [1:5, 1:5] 0 0.129 0.318 0.336 0.751 ...
#> $ p.value.indeptest: num [1:5, 1:5] 0.00 1.53e-05 0.00 0.00 0.00 ...
#> - attr(*, "class")= chr "RVineMatrix"
summary(RVM1)
#> tree edge | family cop par par2 | tau utd ltd
#> ------------------------------------------------------------
#> 1 4,5 | 4 G 3.89 0.00 | 0.74 0.81 -
#> 1,2 | 1 N 0.90 0.00 | 0.72 - -
#> 1,3 | 1 N 0.48 0.00 | 0.32 - -
#> 1,4 | 3 C 4.83 0.00 | 0.71 - 0.87
#> 2 1,5;4 | 4 G 1.49 0.00 | 0.33 0.41 -
#> 4,2;1 | 4 G 1.60 0.00 | 0.37 0.46 -
#> 4,3;1 | 4 G 1.90 0.00 | 0.47 0.56 -
#> 3 3,5;1,4 | 3 C 0.91 0.00 | 0.31 - 0.47
#> 3,2;4,1 | 3 C 1.06 0.00 | 0.35 - 0.52
#> 4 2,5;3,1,4 | 1 N 0.21 0.00 | 0.14 - -
#> ---
#> type: R-vine logLik: 2035.9 AIC: -4051.8 BIC: -4009.66
#> ---
#> 1 <-> V1, 2 <-> V2, 3 <-> V3, 4 <-> V4, 5 <-> V5
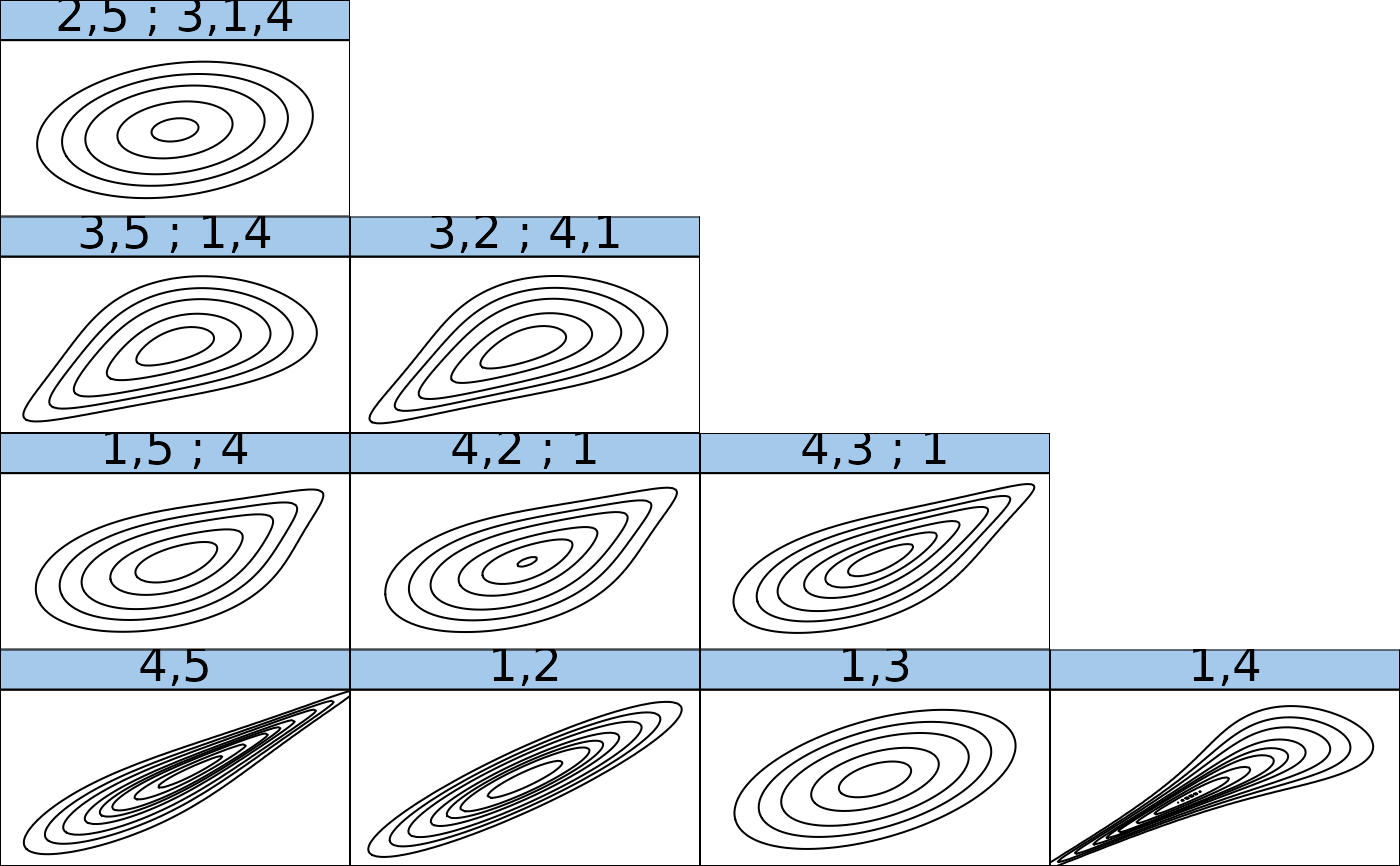
## inspect the fitted model using plots
if (FALSE) plot(RVM1) # tree structure # \dontrun{}
contour(RVM1) # contour plots of all pair-copulas