Simulate from a S-vine model
Arguments
- n
how many steps of the time series to simulate.
- rep
number of replications;
reptime series of lengthnare generated.- model
a S-vine copula model object (inheriting from svinecop_dist).
- past
(optional) matrix of past observations. If provided, time series are simulated conditional on the past.
- qrng
if
TRUE, generates quasi-random numbers using the multivariate Generalized Halton sequence up to dimension 300 and the Generalized Sobol sequence in higher dimensions (defaultqrng = FALSE).- cores
number of cores to use; if larger than one, computations are done parallel over replications.
Value
An n-by-d-byrep array, where d is the cross-sectional
dimension of the model. This reduces to an n-by-d matrix if rep == 1.
Examples
# load data set
data(returns)
returns <- returns[1:100, 1:3]
# fit parametric S-vine model with Markov order 1
fit <- svine(returns, p = 1, family_set = "parametric")
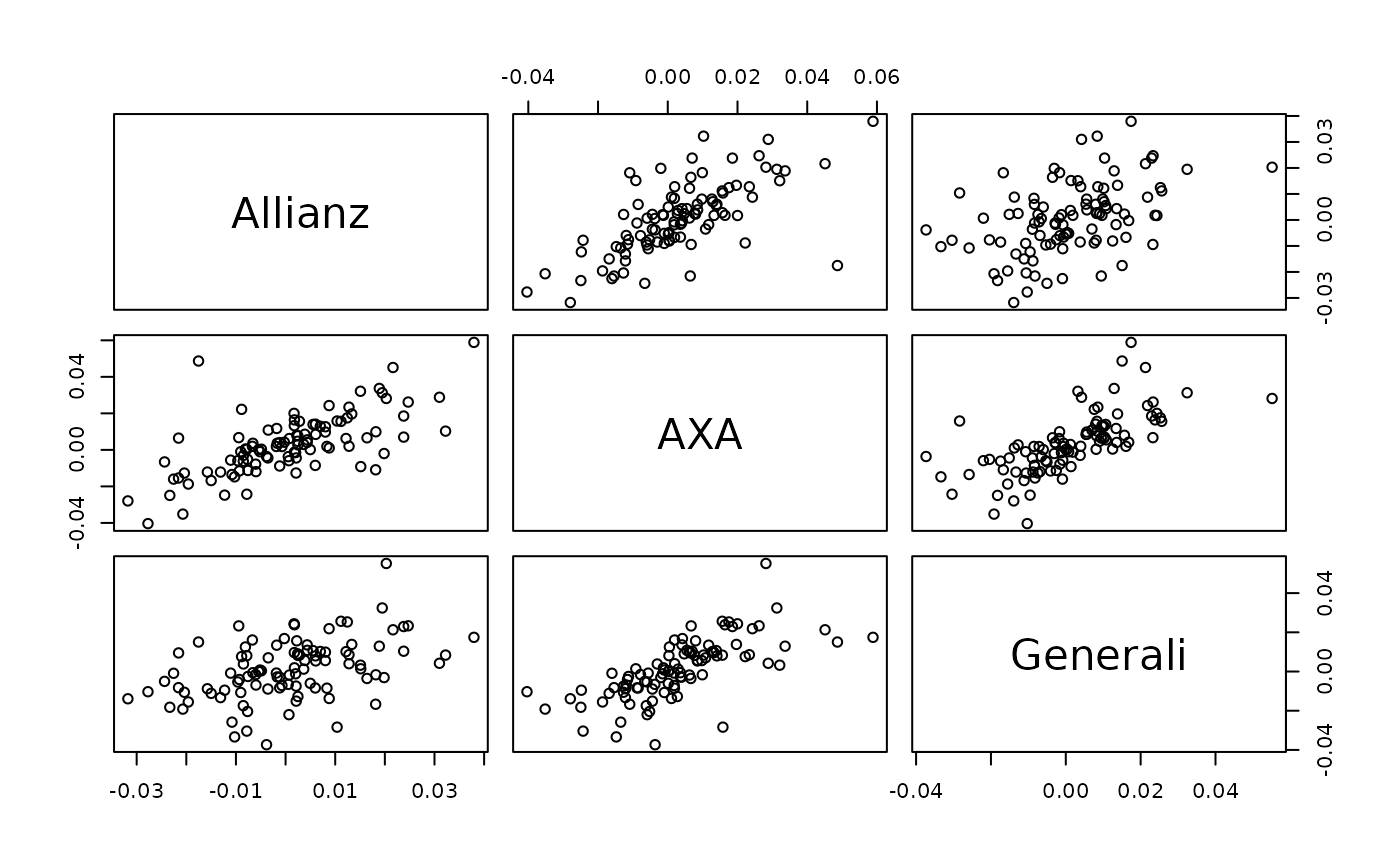
pairs(returns) # original data
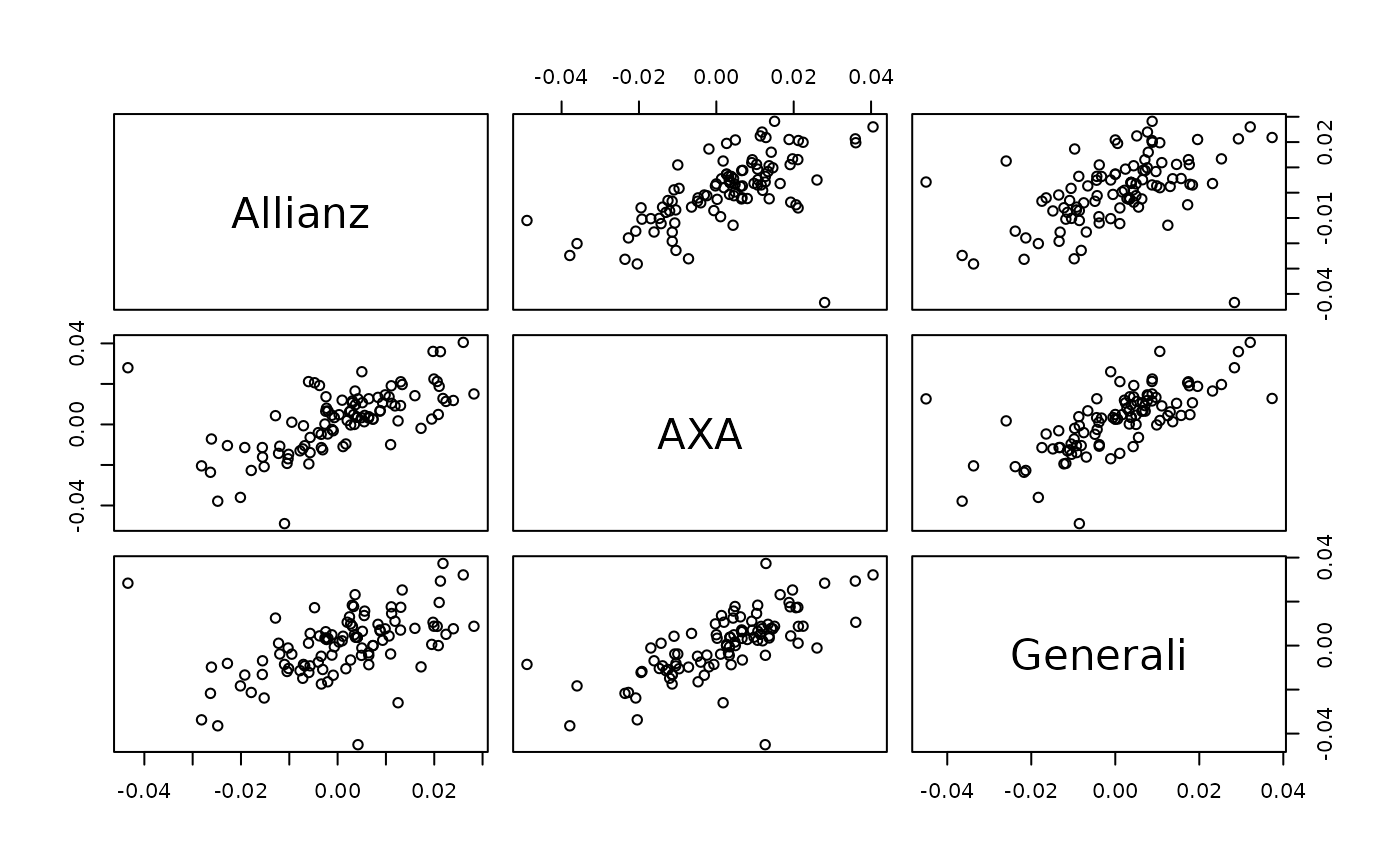 pairs(svine_sim(100, rep = 1, model = fit)) # simulated data
pairs(svine_sim(100, rep = 1, model = fit)) # simulated data
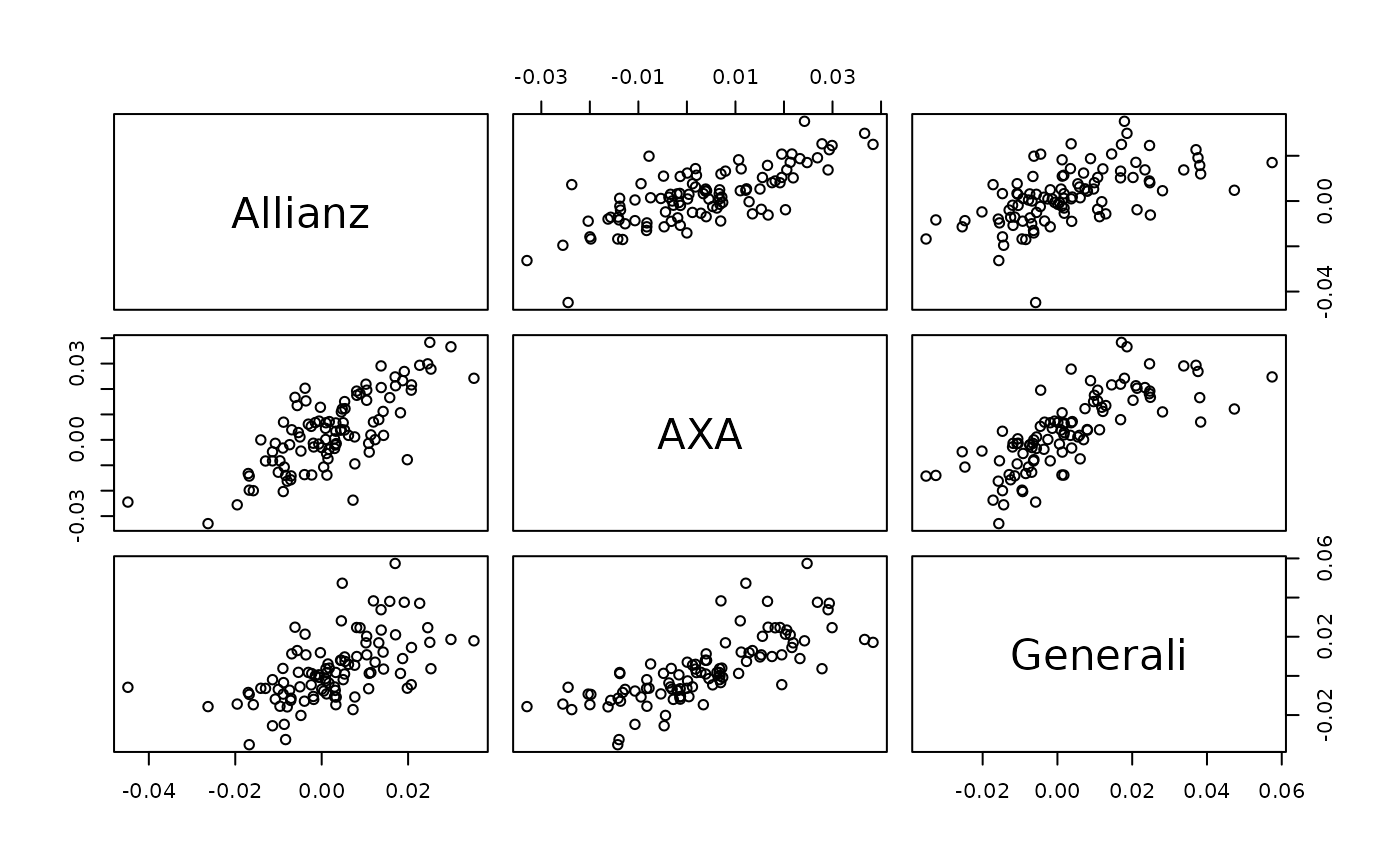 # simulate the next day conditionally on the past 500 times
pairs(t(svine_sim(1, rep = 100, model = fit, past = returns)[1, , ]))
# simulate the next day conditionally on the past 500 times
pairs(t(svine_sim(1, rep = 100, model = fit, past = returns)[1, , ]))