Predict conditional mean and quantiles from a D-vine regression model
Source:R/predict.vinereg.R
predict.vinereg.RdPredict conditional mean and quantiles from a D-vine regression model
Examples
# simulate data
x <- matrix(rnorm(200), 100, 2)
y <- x %*% c(1, -2)
dat <- data.frame(y = y, x = x, z = as.factor(rbinom(100, 2, 0.5)))
# fit vine regression model
(fit <- vinereg(y ~ ., dat))
#> D-vine regression model: y | x.2, x.1
#> nobs = 100, edf = 4, cll = 84.24, caic = -160.49, cbic = -150.07
# inspect model
summary(fit)
#> var edf cll caic cbic p_value
#> 1 y 0 -225.84803 451.6961 451.6961 NA
#> 2 x.2 1 89.80766 -177.6153 -175.0101 5.880492e-41
#> 3 x.1 3 220.28444 -434.5689 -426.7534 3.602588e-95
plot_effects(fit)
#> `geom_smooth()` using method = 'loess' and formula = 'y ~ x'
 # model predictions
mu_hat <- predict(fit, newdata = dat, alpha = NA) # mean
med_hat <- predict(fit, newdata = dat, alpha = 0.5) # median
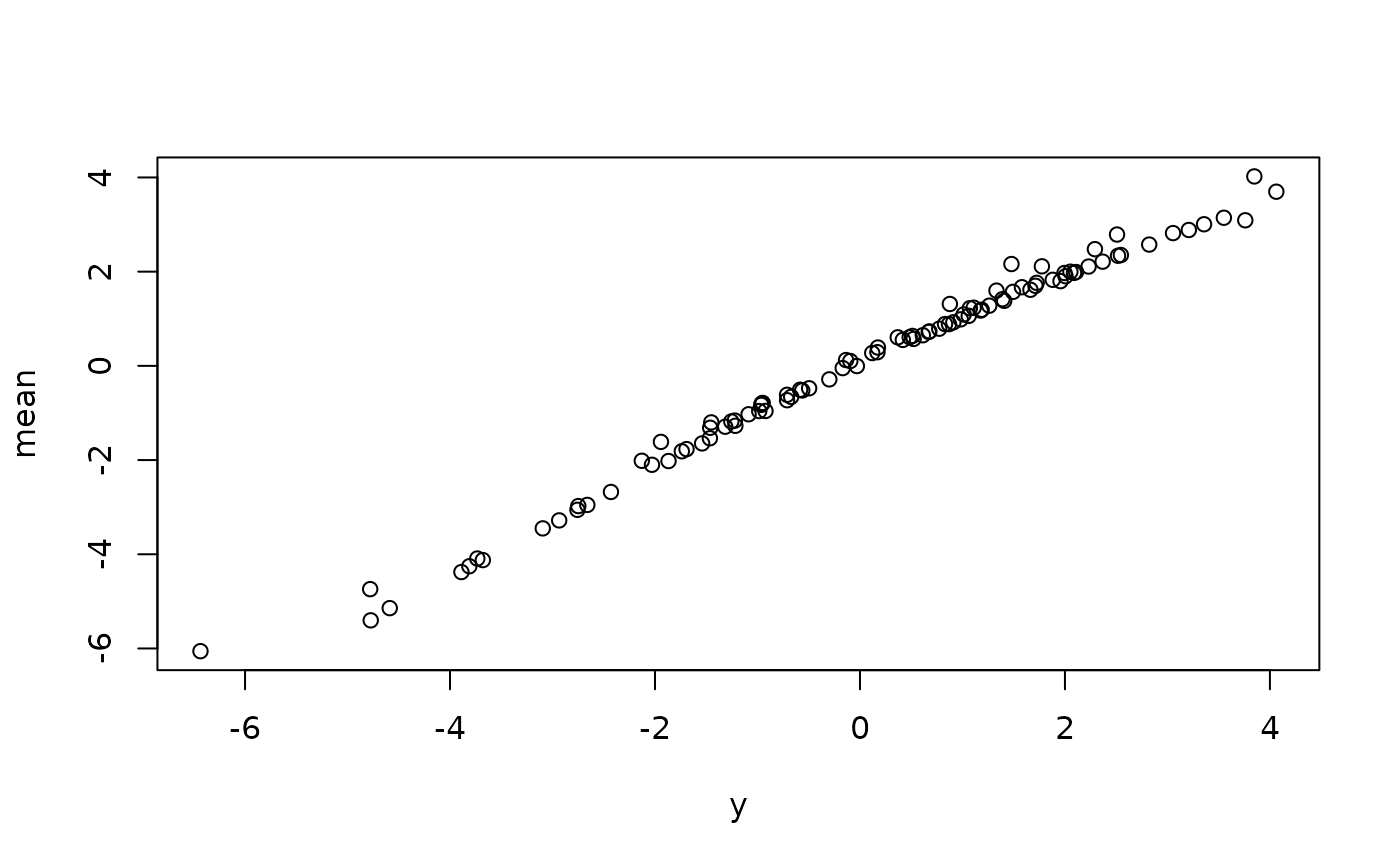
# observed vs predicted
plot(cbind(y, mu_hat))
# model predictions
mu_hat <- predict(fit, newdata = dat, alpha = NA) # mean
med_hat <- predict(fit, newdata = dat, alpha = 0.5) # median
# observed vs predicted
plot(cbind(y, mu_hat))
 ## fixed variable order (no selection)
(fit <- vinereg(y ~ ., dat, order = c("x.2", "x.1", "z.1")))
#> D-vine regression model: y | x.2, x.1, z.1
#> nobs = 100, edf = 4, cll = 84.24, caic = -160.49, cbic = -150.07
## fixed variable order (no selection)
(fit <- vinereg(y ~ ., dat, order = c("x.2", "x.1", "z.1")))
#> D-vine regression model: y | x.2, x.1, z.1
#> nobs = 100, edf = 4, cll = 84.24, caic = -160.49, cbic = -150.07